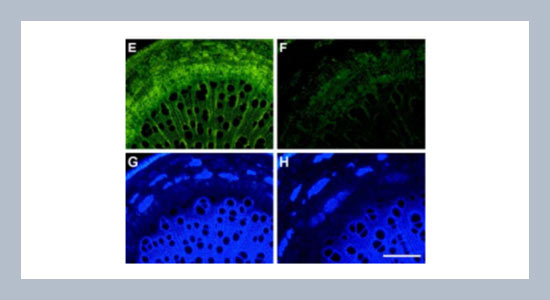
Chung-Jui Tsai*, Walid El Kayal and Scott A. Harding Biotechnology Research Center, School of Forest Resources and Environmental Science,Michigan Technological University,Houghton, MI 49931, U.S.A.
Download Citation:
|
Download PDF
Plant secondary metabolism affects ecosystem diversity and the yield and quality offeedstocks for biomass and biofuel, through an elaborate network of pathways that share commonprecursors. Until recently, functional dissection of these networks has depended largely onmolecular information stored in the genome of Arabidopsis, an annual herb. Now that the Populusgenome sequence is available, the potential for understanding and exploiting secondary metabolismin tree species comes closer to realization. In the present overview, genomic informationpointing to greatly expanded gene complexity and function of the phenylpropanoid pathwayin Populus is summarized. Phenylpropanoid-derived flavonoid and salicylate phenolics occur innumerous functionally distinct forms, and can account for 50% of leaf biomass in Populus andother fast-growing tree taxa. Their potential effects on tree growth, and their documented impactson ecosystem diversity and productivity justify molecular dissection of secondary metabolismin Populus. Biosynthesis of salicylate phenolics remains poorly understood. By contrast, insilico promoter analysis of flavonoid genes, and in situ flavonoid localization in Populus reportedhere, augment published gene expression data, and illustrate that intra and intercellularregulatory components dramatically affect secondary carbon partitioning in this woody perennial. ABSTRACT
Keywords:
lignin; phenolic glycosides; condensed tannins; Populus genome.
Share this article with your colleagues
[1] Croteau, R., Kutchan, T. M., and Lewis,N.G. 2000. Natural products (secondarymetabolites). In: Buchanan B, GruissemW, Jones R, (Eds.) "Biochemistry andMolecular Biology of Plants". AmericanSociety of Plant Physiologists, Rockville,MR: 1250-1318.REFERENCES
[2] Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck, L., Aerts, A., Bhalerao, R. R., Bhalerao, R. P., Blaudez, D., Boerjan, W., Brun, A., Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G. L., Cooper, D., Coutinho, P. M., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve, S., Dejardin, A., dePamphilis, C., Detter, J., Dirks, B., Dubchak, I., Duplessis, S.,Ehlting, J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger, B., Heinze, B., Helariutta, Y., Henrissat, B., Holligan, D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S., Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjarvi, J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri, U., Larimer, F., Leebens-Mack, J., Leple, J.C., Locascio, P., Lou, Y., Lucas, S., Martin,F., Montanini, B., Napoli, C., Nelson, D. R., Nelson, C., Nieminen, K., Nilsson,O., Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J., Richardson, P., Rinaldi, C., Ritland, K.,Rouze, P., Ryaboy, D., Schmutz, J.,Schrader, J., Segerman, B., Shin, H., Siddiqui, A., Sterky, F., Terry, A., Tsai, C.J., Uberbacher, E., Unneberg, P., Vahala, J., Wall, K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., Van, de Peer Y., and Rokhsar, D. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313: 1596-1604.
[3] Boerjan, W., Ralph, J., and Baucher, M.2003. Lignin biosynthesis. Annual Reviewof Plant Biology, 54: 519-546.
[4] Dixon, R. A., Xie, D. Y., and Sharma, S.B. 2005. Proanthocyanidins - a finalfrontier in flavonoid research? NewPhytologist, 165: 9-28.
[5] Winkel-Shirley, B. 2002. Biosynthesis offlavonoids and effects of stress. CurrentOpinion in Plant Biology, 5: 218-223.
[6] Higuchi, T. 1997. "Biochemistry andMolecular Biology of Wood".Springer-Verlag, New York.
[7] Kosbar, L. L., Gelorme, J. D., Japp, R.M., and Fotorny, W. T. 2000. Introducingbiobased materials into the electronicsindustry: Developing a lignin-based resinfor printed wiring boards. Journal of IndustrialEcology, 4: 93-105.
[8] Ghosh, I., Jain, R. K., and Glasser, W. G.1999. Blends of biodegradable thermoplasticswith lignin esters. In: Glasser,W.G., Northey, R.A., Schultz, T.P., (Eds.)"Lignin: Historical, Biological, and MaterialsPrespectives". American ChemicalSociety Series, 742: 331-350.
[9] Kao, Y. Y., Harding, S. A., and Tsai, C. J.2002. Differential expression of two distinctphenylalanine ammonia-lyase genesin condensed tannin-accumulating andlignifying cells of quaking aspen. PlantPhysiology, 130: 796-807.
[10] Hu, W. J., Kawaoka, A., Tsai, C. J., Lung,J. H., Osakabe, K., Ebinuma, H., andChiang, V. L. 1998. Compartmentalizedexpression of two structurally and functionallydistinct 4-coumarate : CoA ligase genes in aspen (Populus tremuloides).Proceedings of the NationalAcademy of Sciences of the United Statesof America, 95: 5407-5412.
[11] Harding, S. A., Leshkevich, J., Chiang, V.L., and Tsai, C. J. 2002. Differential substrateinhibition couples kinetically distinct4-coumarate: coenzyme A ligaseswith spatially distinct metabolic roles inquaking aspen. Plant Physiology, 128:428-438.
[12] Chiang, V. L., and Funaoka, M. 1990.The Difference between guaiacyl andguaiacyl-syringyl lignins in their responsesto Kraft delignification. Holzforschung,44: 309-313.
[13] Li, L., Zhou, Y. H., Cheng, X. F., Sun, J.Y., Marita, J. M., Ralph, J., and Chiang,V. L. 2003. Combinatorial modificationof multiple lignin traits in trees throughmultigene cotransformation. Process NationalAcademic Science U S A, 100:4939-4944.
[14] Hu, W. J., Harding, S. A., Lung, J.,Popko, J. L., Ralph, J., Stokke, D. D.,Tsai, C. J., and Chiang, V. L. 1999. Repressionof lignin biosynthesis promotescellulose accumulation and growth intransgenic trees. Nature Biotechnology,17: 808-812.
[15] Zhong, R., Morrison, W. H., Himmelsbach,D. S., Poole, F. L., and Ye, Z. H.2000. Essential role of caffeoyl coenzymeA O-methyltransferase in ligninbiosynthesis in woody poplar plants.Plant Physiol, 124: 563.
[16] Ralph, J., Lapierre C., Marita, J. M., Kim,H., and Lu, F. 2001. Elucidation of newstructures in lignins of CAD- andCOMT-deficient plants by NMR. Phytochemistry,57: 993.
[17] Baucher, M., Chabbert, B., Pilate, G.,VanDoorsselaere, J., Tollier, M. T., PetitConil,M., Cornu, D., Monties, B.,VanMontagu, M., Inze, D., Jouanin, L.,and Boerjan, W. 1996. Red xylem andhigher lignin extractability bydown-regulating a cinnamyl alcohol dehydrogenasein poplar. Plant Physiol,112: 1479-1490.
[18] Pilate, G., Guiney, E., Holt, K.,Petit-Conil, M., and Lapierre, C. 2002.Field and pulping performances oftransgenic trees with altered lignification.Nature Biotechnology, 20: 607.
[19] Tsai, C. J., Harding, S. A., Tschaplinski,T. J., Lindroth, R. L., and Yuan, Y. 2006.Genome-wide analysis of the structuralgenes regulating defense phenylpropanoidmetabolism in Populus. NewPhytologist, 172: 47-62.
[20] Mahdi, J. G., Mahdi, A. J., Mahdi, A. J.,and Bowen, I. D. 2006. The historicalanalysis of aspirin discovery, its relationto the willow tree and antiproliferativeand anticancer potential. Cell Proliferation,39: 147-155.
[21] Lindroth, R. L., and Hwang, S. Y. 1996.Diversity, redundancy and multiplicity inchemical defense systems of aspen. In:Romeo, J. T., Saunders, J. A., Barbosa,P.,(Eds.) "Recent Advances in Phytochemistry".Plenum Press, New York:25-56.
[22] Warren, J. M., Bassman, J. H., Fellman, J.K., Mattinson, D. S., and Eigenbrode, S.2003. Ultraviolet-B radiation alters phenolicsalicylate and flavonoid compositionof Populus trichocarpa leaves. TreePhysiology, 23: 527-535.
[23] Turtola, S., Rousi, M., Pusenius, J., Yamaji,K., Heiska, S., Tirkkonen, V.,Meier, B., and Julkunen-Tiitto, R. 2005.Clone-specific responses in leaf phenolicsof willows exposed to enhancedUVB radiation and drought stress.Global Change Biology, 11: 1655-1663.
[24] Leon, J., Shulaev, V., Yalpani, N.,Lawton, M. A., and Raskin, I. 1995.Benzoic-acid 2-hydroxylase, a solubleoxygenase from tobacco, catalyzes salicylic-acid biosynthesis. Proceedings ofthe National Academy of Sciences of theUnited States of America, 92:10413-10417.
[25] Wildermuth, M. C., Dewdney, J., Wu, G.,and Ausubel, F. M. 2001. Isochorismatesynthase is required to synthesize salicylicacid for plant defence. Nature, 414:562-565.
[26] Ferrari, S., Plotnikova, J. M., DeLorenzo, G., and Ausubel, F. M. 2003.Arabidopsis local resistance to Botrytiscinerea involves salicylic acid andcamalexin and requires EDS4 and PAD2,but not SID2, EDS5 or PAD4. PlantJournal, 35: 193-205.
[27] Stafford, H. A. 1991. Flavonoid evolution:An enzymic approach. Plant Physiology,96: 680-685.
[28] Winkel-Shirley, B. 2001. FlavonoidBiosynthesis. A Colorful Model for Genetics,Biochemistry, Cell Biology, andBiotechnology. Plant Physiology, 126:485-493.
[29] Zucker, W. V. 1983. Tannins - Doesstructure determine function - an ecologicalperspective. American Naturalist,121: 335-365.
[30] Lindroth, R. L. 1989. Biochemical detoxication- mechanism of differentialtiger swallowtail tolerance to phenolicglycosides. Oecologia, 81: 219-224.
[31] Schimel, J. P., VanCleve, K., Cates, R. G.,Clausen, T. P., and Reichardt, P. B. 1996.Effects of balsam poplar (Populus balsamifera)tannins and low molecularweight phenolics on microbial activity intaiga floodplain soil: Implications forchanges in N cycling during succession.Canadian Journal of Botany, 74: 84-90.
[32] Bradley, R. L., Titus, B. D., and Preston,C. P. 2000. Changes to mineral N cyclingand microbial communities in blackspruce humus after additions of(NH 4 ) 2 SO4 and condensed tannins extractedfrom Kalmia angustifolia andbalsam fir. Soil Biology & Biochemistry,32: 1227-1240.
[33] Lorenz, K., and Preston, C. M. 2002.Characterization of high-tannin fractionsfrom humus by carbon-13cross-polarization and magic-angle spinningnuclear magnetic resonance. Journalof Environmental Quality, 31:431-436.
[34] Stoutjesdijk, P. A., Sale, P. W., andLarkin, P. J. 2001. Possible involvementof condensed tannins in aluminium toleranceof Lotus pedunculatus. AustralianJournal of Plant Physiology, 28:1063-1074.
[35] Davis, M. A., Pritchard, S. G., Boyd, R.S., and Prior, S. A. 2001. Developmentaland induced responses of nickel-basedand organic defences of thenickel-hyperaccumulating shrub, Psychotriadouarrei. New Phytologist, 150:49-58.
[36] Lepiniec, L., Debeaujon, I., Routaboul, J.M., Baudry, A., Pourcel, L., Nesi, N.,and Caboche, M. 2006. Genetics andbiochemistry of seed flavonoids. AnnualReview of Plant Biology, 57: 405-430.
[37] Harborne, J. B. and Williams, C. A. 2000.Advances in flavonoid research since1992. Phytochemistry, 55: 481-504.
[38] Driebe, E. M. and Whitham, T. G. 2000.Cottonwood hybridization affects tanninand nitrogen content of leaf litter and altersdecomposition. Oecologia, 123:99-107.
[39] Whitham, T. G., Young, W. P., Martinsen,G. D., Gehring, C. A., Schweitzer, J. A.,Shuster, S. M., Wimp, G. M., Fischer, D.G., Bailey, J. K., Lindroth, R. L., Woolbright,S., and Kuske C. R. 2003. Communityand ecosystem genetics: A consequenceof the extended phenotype.Ecology, 84: 559-573.
[40] Matsui, K., Tanaka, H., andOhme-Takagi, M. 2004. Suppression ofthe biosynthesis of proanthocyanidin inArabidopsis by a chimeric PAP1 repressor.Plant Biotechnology Journal, 2:487-493.
[41] Tanner, G. J., Francki, K. T., Abrahams,S., Watson, J. M., Larkin, P. J., and Ashton, A. R. 2003. Proanthocyanidinbiosynthesis in plants - Purification oflegume leucoanthocyanidin reductaseand molecular cloning of its cDNA.Journal of Biological Chemistry, 278:31647-31656.
[42] Ayres, M. P., Clausen, T. P., MacLean, S.F., Redman, A. M., and Reichardt, P. B.1997. Diversity of structure and antiherbivoreactivity in condensed tannins.Ecology, 78: 1696-1712.
[43] Veit, M., and Pauli, G. F. 1999. MajorFlavonoids from Arabidopsis thalianaLeaves. Journal of Natural Products, 62:1301-1303.
[44] Prescott, A., Stamford, N., Wheeler, G.,and Firmin, J. 2002. In vitro propertiesof a recombinant flavonol synthase fromArabidopsis thaliana. Phytochemistry, 60:589-593.
[45] Martensa, S. and Mithöfer, A. 2005.Flavones and flavone synthases. Phytochemistry,66: 2399-2407.
[46] Greenaway, W., English, S., Whatley, F.R., and Rood, S. B. 1991. Interrelationshipsof poplars in a hybrid swarm asstudied by gas chromatography-massspectrometry. Canadian Journal of Botany,69: 203-208.
[47] Greenaway, W., English, S., and Whatley,F. R. 1992. Relationships of Populus Xacuminata and Populus X generosa withtheir parental species examined by gaschromatography-mass spectrometry ofbud exudates. Canadian Journal of Botany,70: 212-221.
[48] Maeda, K., Kimura, S., Demura, T., Takeda,J., and Ozeki, Y. 2005. DcMYB1acts as a transcriptional activator of thecarrot phenylalanine ammonia-lyasegene (DcPAL1) in response to elicitortreatment, UV-B irradiation and the dilutioneffect. Plant Molecular Biology, 59:739-752.
[49] Sablowski, R. W. M., Moyano, E., Culianezmacia,F. A., Schuch, W., Martin,C., and Bevan, M. 1994. Aflower-specific Myb protein activatestranscription of phenylpropanoid biosyntheticgenes. Embo Journal, 13:128-137.
[50] Green, P. J., Kay, S. A., and Chua, N. H.1987. Sequence-specific interactions of apea nuclear factor with light-responsiveelements upstream of the Rbcs-3a gene.EMBO Journal, 6: 2543-2549.
[51] Gilmartin, P. M., Sarokin, L., Memelink,J., and Chua, N. H. 1990. Molecular lightswitches for plant genes. Plant Cell, 2:369-378.
[52] Staiger, D., Kaulen, H., and Schell, J.1989. A CACGTG motif of the Antirrhinummajus chalcone synthase promoteris recognized by an evolutionarilyconserved nuclear protein. PNAS, 86:6930-6934.
[53] Takaiwa, F., Yamanouchi, U., Yoshihara,T., Washida, H., Tanabe, F., Kato, A., andYamada, K. 1996. Characterization ofcommon cis-regulatory elements responsiblefor the endosperm-specific expressionof members of the rice glutelin multigenefamily. Plant Molecular Biology,30: 1207-1221.
[54] Suzuki, A., Wu, C. Y., Washida, H., andTakaiwa, F. 1998. Rice MYB proteinOSMYB5 specifically binds to theAACA motif conserved among promotersof genes for storage protein glutelin.Plant and Cell Physiology, 39: 555-559.
[55] Debeaujon, I., Nesi, N., Perez, P., Devic,M., Grandjean, O., Caboche, M., andLepiniec, L. 2003. Proanthocyanidin-accumulating cells in Arabidopsistesta: Regulation of differentiation androle in seed development. Plant Cell, 15:2514-2531.
[56] Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K.,Umemura, I., Hasegawa, Y., Ashikari, M.,Kitano, H., and Matsuoka, M. 2005.Crown rootless1, which is essential forcrown root formation in rice, is a targetof an AUXIN RESPONSE FACTOR in:auxin signaling. Plant Cell, 17:1387-1396.
[57] Fujita, Y., Fujita, M., Satoh, R., Maruyama,K., Parvez, M. M., Seki, M., Hiratsu,K., Ohme-Takagi, M., Shinozaki,K., and Yamaguchi-Shinozaki, K. 2005.AREB1 is a transcription activator ofnovel ABRE-dependent ABA signalingthat enhances drought stress tolerance inArabidopsis. Plant Cell, 17: 3470-3488.
[58] Choi, H. I., Hong, J. H., Ha, J. O., Kang,J. Y., and Kim, S. Y. 2000. ABFs, a familyof ABA-responsive element bindingfactors. Journal of Biological Chemistry,275: 1723-1730.
[59] Block, A., Dangl, J. L., Hahlbrock, K.,and Schulze-Lefert, P. 1990. Functionalborders, genetic fine structure, and distancerequirements of cis elements mediatinglight responsiveness of the parsleychalcone synthase promoter. PNAS, 87:5387-5391.
[60] Feucht, W., and Treutter, D. 1990. Flavan-3-ols in trichomes, pistils and phellodermof some tree species. Annals ofBotany, 65: 225-230.
[61] Neu, R. 1956. A new reagent for differentiatingand determining flavones onpaper chromatograms. Naturwissenschaften,43: 82.
[62] Harding, S. A., Jiang, H. Y., Jeong, M. L.,Casado, F. L., Lin, H. W., and Tsai, C. J.2005. Functional genomics analysis offoliar condensed tannin and phenolicglycoside regulation in natural cottonwoodhybrids. Tree Physiology, 25:1475-1486.
ARTICLE INFORMATION
Available Online:
2006-12-03
Tsai, C.-J., Kayal, W.-E., Harding, S.-A. 2006. Populus, the new model system for investigating Phenyl-propanoid complexity. International Journal of Applied Science and Engineering, 4, 221–233. https://doi.org/10.6703/IJASE.2006.4(3).221
Cite this article: