Ganesh Prasad Mishra1*, Debadash Panigrahi2 1 Kharvel Subharti College of Pharmacy, Swami Vivekanand Subharti University, Subhartipuram, Meerut, U.P, India
2 Drug Research Laboratory, Nodal Research Centre, College of Pharmaceutical Sciences, Puri, Odisha, India
Download Citation:
|
Download PDF
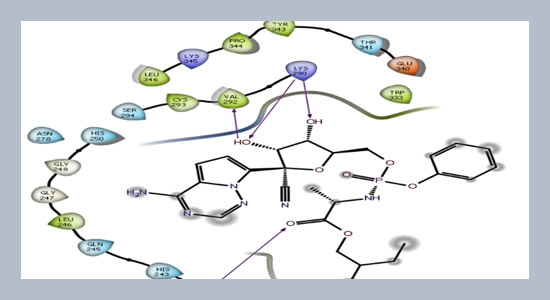
The recent pandemic caused by SARS-CoV-2 is a major health concern for the entire world. Fast transmission and the mortality rate of this disease is a serious threat to human society. By looking into its seriousness finding medication against this disease is the biggest task of the present time. In the present study, computational approach is used to find effective compounds against SARS-CoV-2 infections. 1452 compounds having potential antiviral activity were selected from PubChem database to search potential inhibitors for the therapeutic targets of SARS-CoV-2. Molecular docking and absorption, distribution, metabolism, elimination and toxicity (ADMET) properties study were performed using Maestro 12.4 (Schrodinger Suite). All the selected compounds were funneled down to six compounds which are found to be effective against all the five targets of SARS-CoV-2. Among six compounds, compound C6 is exhibited excellent docking score -8.93, -8.21, -7.93, -6.73 kcal/mol with main protease (Mpro), angiotensin-converting enzyme-2 (ACE2), RNA dependent RNA polymerase (RdRp), and endoribonuclease (NSP15) target proteins respectively.ABSTRACT
Keywords:
Molecular docking, Computational screening, ADMET, COVID-19, Multi-target, SARS-CoV-2, Antiviral compounds.
Share this article with your colleagues
REFERENCES
ARTICLE INFORMATION
Received:
2020-10-27
Accepted:
2020-12-31
Available Online:
2021-06-01
Mishra, G.P., Panigrahi, D. 2021. Molecular docking and ADMET study for searching multi targeted antiviral compounds against SARS-CoV-2: A computational approach, International Journal of Applied Science and Engineering, 18, 2020273. https://doi.org/10.6703/IJASE.202106_18(2).004
Cite this article:
Copyright The Author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are cited.